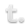Category Archives: news
New pre-print: GCAD: a Computational Framework for Mammalian Genetic Program Computer-Aided Design
We’re excited to share our new pre-print which develops a computational framework to aid in mammalian genetic circuit design. Congrats to Katie Dreyer and the whole GCAD team! Check out the pre-print here!
New paper: Conversion of natural cytokine receptors into orthogonal synthetic biosensors
We’re excited to share our latest work in which we coopt naturally evolved receptor-ligand interactions and integrate them with self-contained signaling modules to build synthetic receptors that are well-suited to creating customized cellular therapies. Congratulations to Dr. Hailey Edelstein and Dr. Amparo Cosio. Check out the published paper here and the preprint here!
New pre-print: Distinguishing Protein and Gene Delivery Enables Characterization and Bioengineering of Extracellular Vesicle-Adeno-Associated Virus Vectors
We’re excited to share our new pre-print which develops an assay which distinguishes protein vs. gene delivery to examine effects of surface-modification on extracellular vesicle encapsulated adeno-associated virus (EV-AAV)-mediated gene delivery. Congrats to Jonathan Boucher and the whole EV-AAV team! Check out the pre-print here!

New paper: Engineered Feedback Employing Natural Hypoxia-Responsive Factors Enhances Synthetic Hypoxia Biosensors
We’re excited to share our new paper which uses mathematical modeling to enhance the performance of a hypoxia sensing biosensor through feedback regulation. Congrats to Dr. Katie Dreyer, Dr. Patrick Donahue, and the whole HBS team ACS SynBio! Check out the paper here!
Amparo Cosio successfully defends her Ph.D. dissertation. Congratulations, Dr. Cosio!
Will Corcoran successfully defends his Ph.D. dissertation. Congratulations, Dr. Corcoran!
New paper: Exploring Structure—Function Relationships in Engineered Receptor Performance Using Computational Structure Prediction
We’re excited to share our recent work examining how protein structure prediction tools can be used to explore structure-function relationships in synthetic receptors. Our work was featured on the cover of GEN Biotechnology Vol. 4, No. 1. Congratulations to Dr. Will Corcoran, Amparo Cosio, and our alumni Dr. Hailey Edelstein. Check out the paper here!
New paper: Developing, characterizing, and modeling CRISPR-based point-of-use pathogen diagnostics
We’re excited to share our recent work developing a nucleic acid sequence-based amplification pathogen diagnostic for SARS-CoV-2, with 20-200 aM sensitivity. We also develop an explanatory model to gain mechanistic insight into reaction performance. Congratulations to Dr. Katie Dreyer, Dr. Kate Dray, and our wonderful collaborators in the Lucks, Jewett, and Mangan groups. Check out the paper here!
New preprint: Exploring structure-function relationships in engineered receptor performance using computational structure prediction
We’re excited to share our recent work discovering how structure influences synthetic receptor function—our lab’s first study using protein structure prediction! Congratulations to Will, Amparo, and Dr. Hailey Edelstein. Check out the preprint here!
Katie Dreyer successfully defends her Ph.D. dissertation. Congratulations, Dr. Dreyer!
New preprint: Engineered feedback employing natural hypoxia-responsive factors enhances synthetic hypoxia biosensors
We’re thrilled to share our recent work building improved hypoxia biosensors using genetic circuits, and improving understanding of the hypoxia response by developing a mathematical model for the biosensors. Congratulations to Katie, Dr. Patrick Donahue, and the rest of the team. Check out the preprint here!
New preprint: Developing, characterizing and modeling CRISPR-based point-of-use pathogen diagnostics
We’re excited to share our recent work developing a nucleic acid sequence-based amplification pathogen diagnostic for SARS-CoV-2, with 20-200 aM sensitivity. We also develop an explanatory model to gain mechanistic insight into reaction performance. Congratulations to Katie, Dr. Kate Dray, and our wonderful collaborators in the Lucks, Jewett, and Mangan groups. Check out the preprint here!
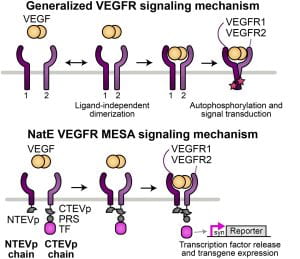
New paper: HaloTag display enables quantitative single-particle characterisation and functionalisation of engineered extracellular vesicles
We’re excited to announce our recent work developing a method using flexible HaloTag labeling to absolutely quantify surface protein loading at the individual EV level, providing new insights into EV heterogeneity. An interesting new insight is that existing methods can substantially underestimate surface display. Congratulations to Dr. Roxi Mitrut, Dr. Devin Stranford, and Beth. Check out the paper here!
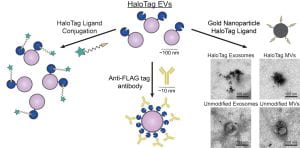
New paper: Enhancing extracellular vesicle cargo loading and functional delivery by engineering protein-lipid interactions
We’re excited to share our recent collaboration with the Kamat Lab, exploring how lipid-protein interactions may be leveraged to direct proteins into extracellular vesicles. Congratulations Dr. Taylor Gunnels, Dr. Hailey Edelstein, and our excellent collaborators Dr. Justin Peruzzi and Dr. Neha Kamat of the Kamat Lab. Check out the paper here and the press piece here!
Iva Hammitt-Kess successfully defends her MS dissertation. Congratulations, Iva!
Taylor Gunnels successfully defends his Ph.D. dissertation. Congratulations, Dr. Gunnels!
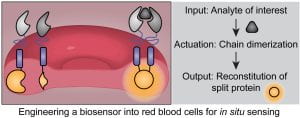
New paper: Building Synthetic Biosensors Using Red Blood Cell Proteins
We’re excited to share our recent work developing genetically-encoded biosensors for use in red blood cells and validating their function in living animals. Congratulations to Dr. Taylor Dolberg and Taylor Gunnels. Check out the paper here!
Roxi Mitrut successfully defends her Ph.D. dissertation. Congratulations, Dr. Mitrut!
New paper: Genetically encoding multiple functionalities into extracellular vesicles for the targeted delivery of biologics to T cells
We’re very excited to share our latest work, now published in Nature Biomedical Engineering! Here, we introduce the GEMINI platform for engineering multifunctional extracellular vesicles as versatile, programmable, biological delivery vehicles. Congratulations to Dr. Devin Stranford and Beth. Check out the paper here and check out a press article about the paper here!
New preprint: HaloTag display enables quantitative single-particle characterization and functionalization of engineered extracellular vesicles
We’re excited to announce our recent work developing a method using flexible HaloTag labeling to absolutely quantify surface protein loading at the individual EV level, providing new insights into EV heterogeneity. An interesting new insight is that existing methods can substantially underestimate surface display. Congratulations to Roxi and Devin. Check out the preprint here!
Simrita Deol successfully defends her Ph.D. dissertation. Congratulations, Dr. Deol!
New Paper: Teaching systematic, reproducible model development using synthetic biology
We’re excited to share our recent work developing a resource for teaching mathematical model development in synthetic biology, with associated open-source teaching materials. The teaching module makes use of our previously published GAMES tutorial. Congratulations to Dr. Kate Dray and Katie, as well as our collaborator, Dr. Julius Lucks. Check out the paper here!
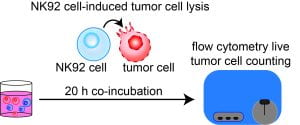
New paper: Comparative evaluation of synthetic cytokines for enhancing production and performance of NK92 cell-based therapies
We’re excited to share our latest work focused on evaluating synthetic cytokines to enhance the production and performance of promising off-the-shelf cell therapies for treating cancer. We engineered natural killer-like NK92 cells to express six synthetic cytokines and evaluated their utility for driving expansion, cancer cell killing, and resistance to tumor microenvironmental conditions. Congratulations to Simrita, Patrick, Roxi, and Iva. Check out the paper here!
New preprint: Enhancing extracellular vesicle cargo loading and functional delivery by engineering protein-lipid interactions
We’re excited to share our recent collaboration with the Kamat Lab, exploring how lipid-protein interactions may be leveraged to direct proteins into extracellular vesicles. Congratulations Taylor, Hailey, and our excellent collaborators Justin and Dr. Neha Kamat of the Kamat Lab. Check out the preprint here!
Hailey Edelstein successfully defends her Ph.D. dissertation. Congratulations Dr. Edelstein!
New review article: The sound of silence: Transgene silencing in mammalian cell engineering
We’re excited to announce the publication of a comprehensive review on the challenges and opportunities for modulating transgene silencing in mammalian cells! Congratulations to Hailey, who co-lead this important work in collaboration with many leaders of the mammalian synthetic biology community. Check out the paper here!
Congratulations to Dr. Devin Stranford for being selected to receive the 2022 International Institute for Nanotechnology Outstanding Researcher Award!
Way to go, Devin!
Kate Dray successfully defends her Ph.D. dissertation. Congratulations Dr. Dray!
Special acknowledgements to science dog Melvin!
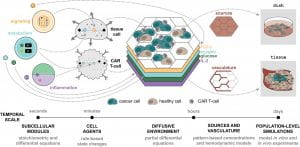
New paper: Mapping CAR T-cell design space using agent-based model
We’re excited to announce our latest work, now published in Frontiers in Molecular Biosciences! Through a collaboration with the Bagheri lab, we developed an agent-based computational testbed to explore how tunable CAR T-cell features and tumor properties impact solid tumor treatment. We adapted the Bagheri lab’s ARCADE framework to develop CAR T-cell Agent-based Representation of Cells And Dynamic Environments (CARCADE). This model enables one to explore how changing key tumor/treatment features impacts therapeutic efficacy, yielding hypotheses that correspond to various experimental equivalents. In this study, we validate the model, interrogate the biological underpinnings of various treatment outcomes, and propose novel tuned treatment strategies that can be tested experimentally. Congratulations to Alex and our excellent collaborators Dr. Jessica Yu and Dr. Neda Bagheri of the Bagheri lab! Check out the paper here, and access the CARCADE code freely on GitHub here!
Dr. Alex Prybutok awarded ChBE Distinguished Trainee Service Award
New preprint: Bioengineering multifunctional extracellular vesicles for targeted delivery of biologics to T cells
We’re very excited to share our recent EV engineering work, now on bioRxiv! Here, we introduce the GEMINI platform for engineering multifunctional extracellular vesicles as programmable biological delivery vehicles. We demonstrate EV targeting, cargo loading, and fusion capabilities by delivering Cas9-sgRNA complexes to primary human T cells to knock out an HIV co-receptor. Check out the preprint here!
Undergraduate Yazeed Alroogi awarded Michael Jaharis Undergraduate Research Fellowship
Alex Prybutok successfully defends her Ph.D. dissertation. Congrats Dr. Prybutok!
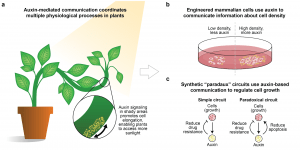
New perspective: Engineering Mammalian Cells to Communicate Using a Language from Plants
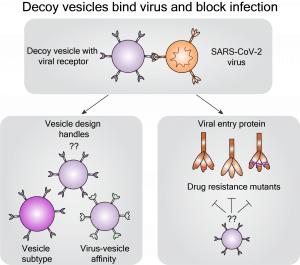
New paper: Elucidating Design Principles for Engineering Cell-Derived Vesicles to Inhibit SARS-CoV-2 Infection
We’re excited to share that our study investigating nanoscale decoys as SARS-CoV-2 inhibitors is out in Small! We learned that engineered decoy nanoparticles are potent viral inhibitors and appear quite resistant to viral evolutionary escape. We hope that the design rules and performance metrics we identified will accelerate the development and preclinical evaluation of this exciting new class of antiviral therapeutics. Congratulations to Taylor, Devin, and Roxi and our excellent collaborator Dr. Neha Kamat. Check out the paper here! Additionally, check out some press about the paper here!
Undergraduates Iva Hammitt-Kess and Alex Beres awarded Summer Undergraduate Research Grants
Beth DiBiase awarded NSF Graduate Research Fellowship
New review article: The evolution of synthetic receptor systems
We’re thrilled to announce the publication of a review in which we survey the existing mammalian synthetic biology toolkit of protein-based receptors and signal-processing components, highlighting efforts to evolve and integrate some of the foundational synthetic receptor systems. This is the review we wish existed, so we wrote it ourselves! Congratulations to Hailey, and our excellent collaborators Janvie and Dr. Leonardo Morsut. Check out the paper here!
New paper: GAMES: A dynamic model development workflow for rigorous characterization of synthetic genetic systems.
We’re thrilled to announce the publication of GAMES, a model development workflow for rigorous characterization of synthetic genetic systems. Congratulations to Kate and Joe, and our excellent collaborators Dr. Niall Mangan and Dr. Neda Bagheri. We hope that GAMES will be useful for synthetic and systems biology researchers interested in systematic, reproducible dynamic model development. Check out the paper here!
New preprint: Elucidating design principles for engineering cell-derived vesicles to inhibit SARS-CoV-2 infection
We’re excited to introduce our work demonstrating that biologics called “decoy” vesicles can inhibit SARS-CoV-2 infection and are robust to multiple problematic viral mutations. Congratulations to Taylor and Devin, and our excellent collaborator Dr. Neha Kamat. We hope that the engineering insights identified in this study will accelerate the development of this exciting new class of antiviral therapeutics (for COVID-19 and/or other diseases). Check out the preprint here!
New preprint: GAMES: A dynamic model development workflow for rigorous characterization of synthetic genetic systems.
We’re thrilled to introduce GAMES, a model development workflow for rigorous characterization of synthetic genetic systems. Congratulations to Kate and Joe, and our excellent collaborators Dr. Niall Mangan and Dr. Neda Bagheri. We hope that GAMES will be useful for synthetic and systems biology researchers interested in systematic, reproducible dynamic model development. Check out the preprint here!
Hailey Edelstein wins 3rd Place poster prize at the Central US Synthetic Biology Workshop
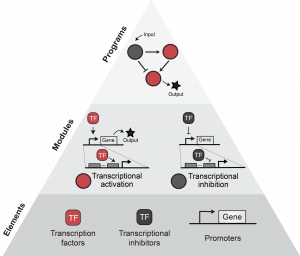
New Paper: Control of mammalian cell-based devices with genetic programming
We’re excited to announce that our new review on the power of using process control concepts to understand and build genetic programs in mammalian cells is out in Current Opinions in Systems Biology! Congrats to Kate, Hailey, and Katie on this informative review (with beautiful figures!)! Check out the new paper here!
Gauri Bora awarded Synthesizing Biology Across Scales National Research Traineeship Program fellowship
Roxi Mitrut awarded Biotechnology Training Program fellowship
Amparo Cosio awarded Biotechnology Training Program fellowship
Alex Prybutok awarded Terminal Year Fellowship
Devin Stranford successfully defends her Ph.D. dissertation. Congrats Dr. Stranford!
Erin awarded Northwestern Summer Undergraduate Research Grant
Taylor Gunnels awarded NSF Graduate Research Fellowship
New Paper: Model-guided design of mammalian genetic programs is out in Science Advances
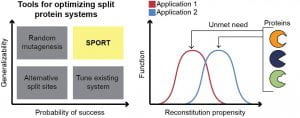
New paper: Computation-guided optimization of split protein systems is out in Nature Chemical Biology
We’re excited that SPORT, a computational protein design strategy for adjusting reconstitution propensity to fall into the sweet spot for your application, has officially been released in Nature Chemical Biology today. This Rosetta-based pipeline identifies mutations that tweak reconstitution propensity so that a split protein comes together only when needed – a property that is useful for creating protein-based sensors, biochemical assays, and parts for synthetic biology. Congratulations to Taylor, Jon, Will, Elizabeth, Alex, and our excellent and key collaborators Srivatsan Raman and Tony Meger on this fantastic effort! Check out our new paper here! Additionally, check out some press from Northwestern Engineering on the study here!
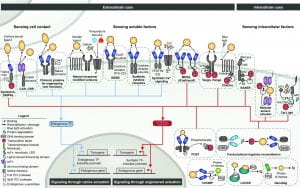
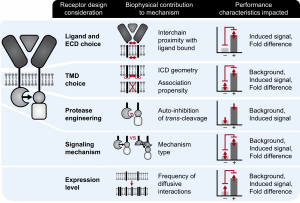
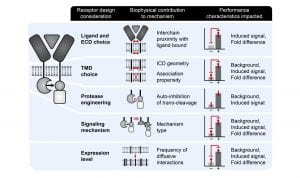
New paper: Elucidation and refinement of synthetic receptor mechanisms is out in Synthetic Biology
We are thrilled to announce our paper synthetic receptor mechanisms is out in Synthetic Biology today! In this study, took a deep dive into Modular Extracellular Sensor Architecture (MESA) design, identified several handles for substantially improving performance, and applied these findings to build next-gen receptors. We hope that these insights will help guide the use of MESA receptors for many applications and may also inform the design and refinement of other synthetic receptors. Congrats to Hailey, Patrick, Joe, Anthony, Taylor, Lauren, Everett, and Amy! Check out our paper here!
Patrick Donahue successfully defends his Ph.D. dissertation. Congrats Dr. Donahue!
Josh and Michelle’s patent of EV targeted was highlighted by Nature Biotechnology
You can read more about the highlight here.
Katie Dreyer awarded a fellowship from the Doctoral Interdisciplinary Cluster on Predictive Science and Engineering Design (PS&ED)
Joe Muldoon wins IBiS Outstanding Thesis Prize
Jon Boucher received the 2019-2020 IBiS Outstanding TA Award
Patrick Donahue receives Rappaport Award for Research Excellence
Joe Muldoon successfully defends his Ph.D. dissertation. Congrats Dr. Muldoon!
Devin Stranford awarded Terminal Year Fellowship
Anthony Kang awarded the Irving M. Klotz Prize for Basic Research for his outstanding honors thesis
Taylor Dolberg successfully defends her Ph.D. dissertation. Congrats Dr. Dolberg!
Quorum Licensing paper highlighted in Science
Science highlights the quorum licensing paper discussing the “wisdom of the crowd” strategy used by cells to make decisions as a population in an article covering highlights from papers published in other journals.
You can the piece here.
Kate featured on the Northwestern International Institute for Nanotechnology’s website
Kate was interviewed by the Northwestern International Institute for Nanotechnology. The interview covers a range of topics, including what got her interested in engineering, her research interests, and her personal challenges and highlights from Northwestern! Thanks for sharing your important and deeply personal story and inspiring others, Kate!
Read the full interview here.
Liz awarded the Gotaas Award, Northwestern’s McCormick School of Engineering’s highest award for undergraduate research
Joe featured on the Northwestern International Institute for Nanotechnology’s website
Joe was interviewed by the Northwestern International Institute for Nanotechnology. The interview covers a range of topics, including what got him interested in engineering, his research interests, his hobbies outside of work, and highlights from his time here at Northwestern!
Read the full interview here.
Liz and Anthony invited to give talks at the Northwestern Undergraduate Research Expo
Austin awarded Northwestern Summer Undergraduate Research Grant
NU SynBio teams up to fight coronavirus and award NSF RAPID grant
In a powerful collaboration between the Leonard, Lucks, Mangan, and Jewett labs, NU SynBio teams up to create a one-step diagnostic tool, and recently received funding for the effort. Congrats to the whole team on this massive effort to aid in the current pandemic!
New pre-print: Elucidation and refinement of synthetic receptor mechanisms
We’re thrilled to share our new paper studying next gen synthetic receptors with improved performance and expanded sensing options by taking cues from nature (and good old biophysics). Congratulations to , , , , , , , and Amy! Check out our pre-print here.
Former Leonard Lab REU undergraduate and senior at Georgia Tech Katie Zhu awarded Fulbright Fellowship
Jon awarded NSF Graduate Research Fellowship and Katie awarded NSF GRFP Honorable Mention
COMET paper highlighted in Northwestern press piece
Check out the press piece, published by Northwestern’s McCormick School of Engineering, on COMET, our new platform for composing genetic programs in mammalian cells!
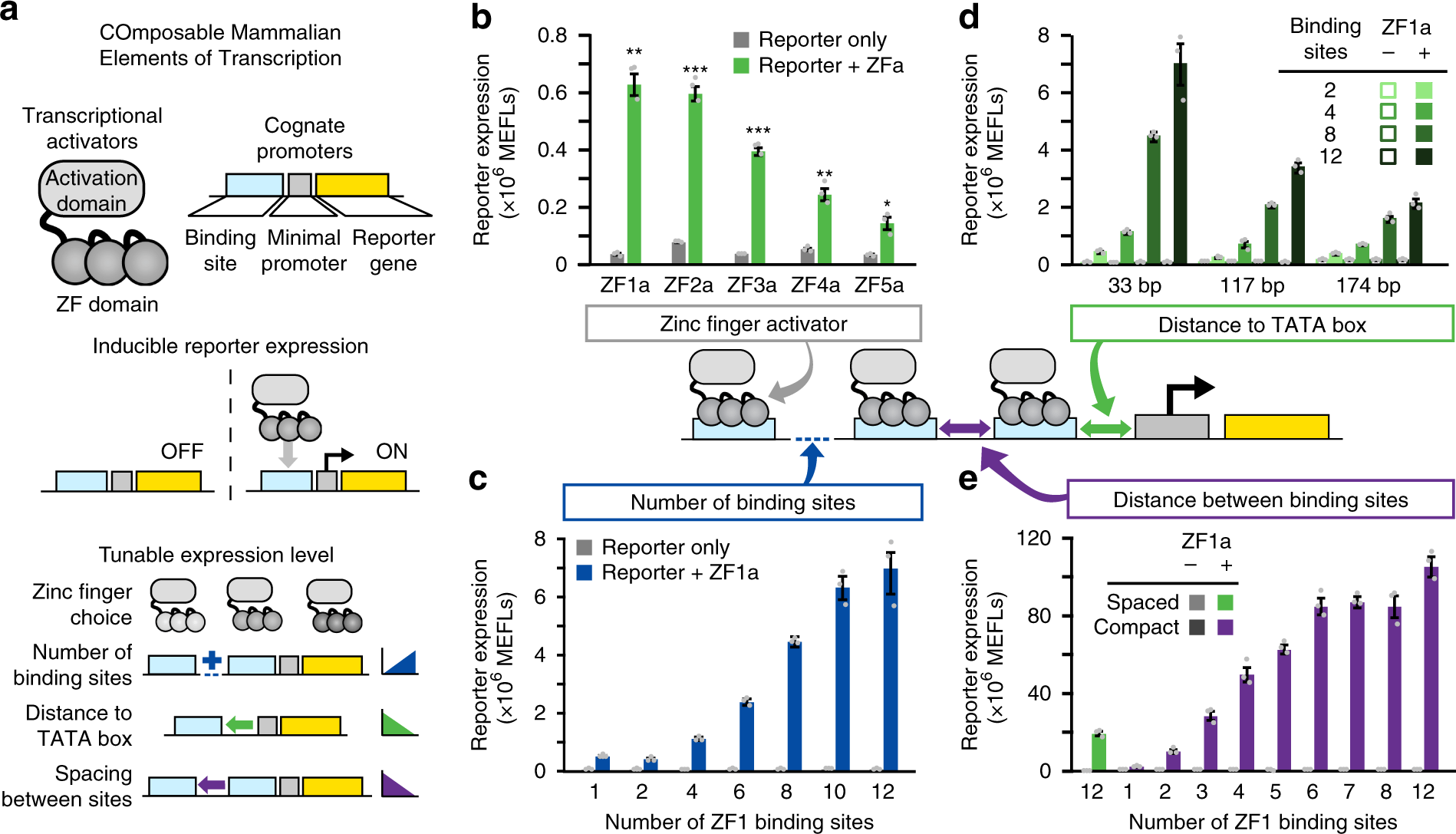
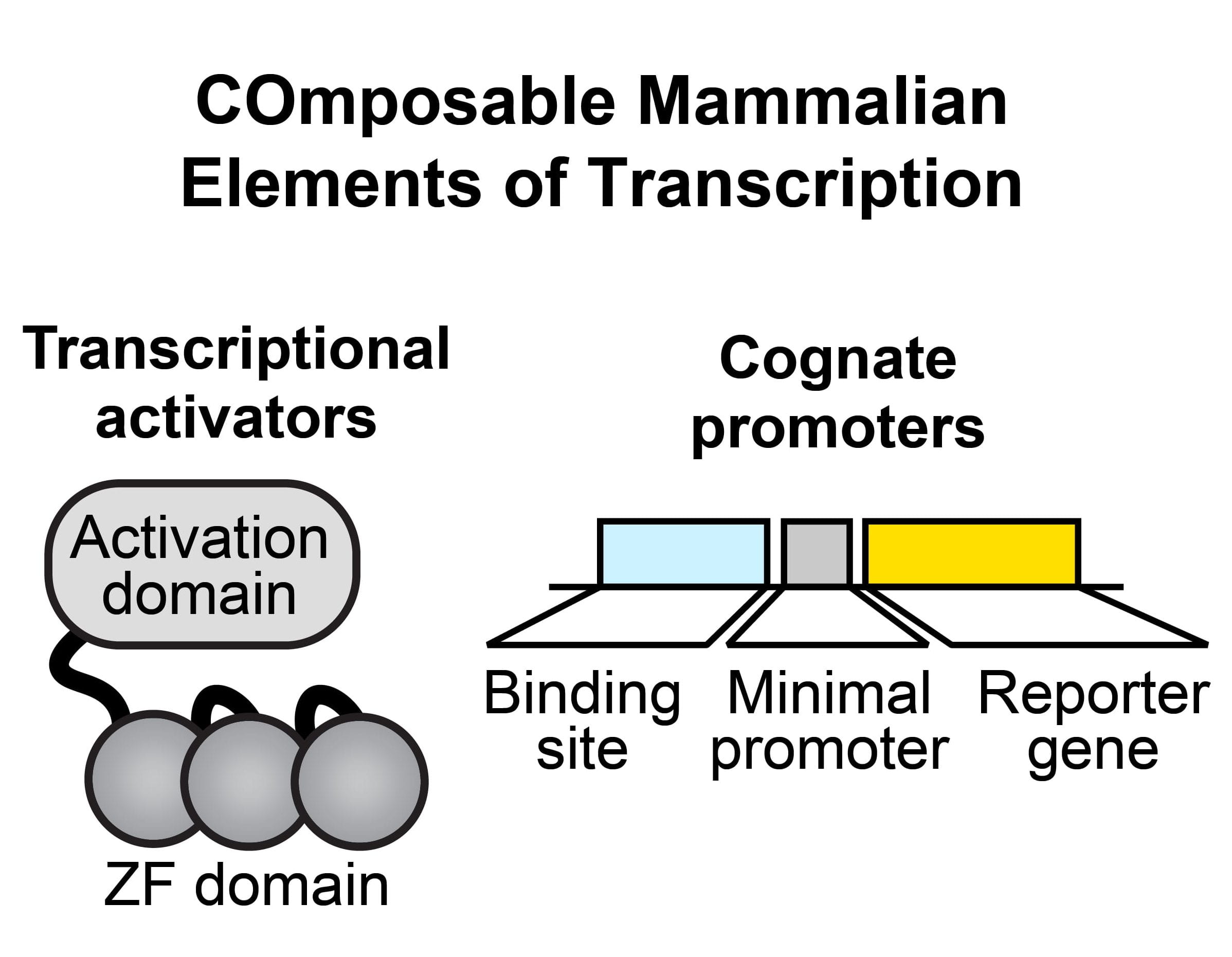
New paper: COMET the toolkit for programming mammalian gene expression is out in Nature Communications
New preprint: Computation-guided optimization of split protein systems
We’re thrilled to introduce SPORT, a computational protein design strategy for adjusting reconstitution propensity to fall into the sweet spot for your application. Congratulations to Taylor, Jon, Will, Elizabeth, Alex, and our excellent and key collaborators Srivatsan Raman and Tony Meger. This exciting and collaborative work involved a lot of hard work on both the experimental and computational efforts, and is an exciting step in split protein optimization for a wide array of applications! Check out our preprint here!
Center for Synthetic Biology Faculty work with Artist-at-Large Dario Robleto to think about how their research affects society and are featured in a video about the collaboration
Dario Robleto returns to Northwestern as the new Artist-at-Large, working with faculty all over campus to discuss the implications of their research on society and ethics. Several professors in the Center for Synthetic Biology, including Josh Leonard, Danielle Tullman-Ercek, and Julius Lucks, are featured in a video about the collaboration.
To learn more about this exciting, and thought provoking collaboration, check out the video here!
Anthony presents his summer research at the AOSC poster session
Josh named a 2019 Researcher to Know
New pre-print: the COMET mammalian synthetic biology toolkit
We’re thrilled to share a new toolkit for mammalian synthetic biology: the COMET library of synthetic transcription factors, promoters, and accompanying models. Congratulations to Patrick, Joseph, Joe and Hailey and our key collaborator Neda Bagheri! This is true tour-de-force many years in the making, and we’re excited to share this with the community! Check out our preprint here, and follow some twitter discussion here.
Will Corcoran awarded Biotechnology Training Program Cluster fellowship
Jon Boucher awarded Biotechnology Training Program fellowship
Josh featured in article commenting on Fussenegger’s mint-triggered gene switches
Josh Leonard’s commentary was featured in a news article in Chemical & Engineering News in July featuring synthetic biologist Martin Fusseneger’s menthol-triggered insulin-producing cells. Josh commented saying the work is “an important first step”, as it was the first time a gene circuit was made entirely from human parts.
Find the full news article here.
Congrats to recent graduates!
Liz wins Audience Choice Award for her poster presentation at the Undergraduate Research and Arts Exposition
Vis awarded the Biomedical Engineering Department Award for Research
Josh Leonard participates in interdisciplinary ethics panel at The Block Museum of Art
The Block Museum of Art hosted “Exploring Ethics: Across Art, Humanities, and Science”, a public event that provided researchers in various fields and artists a platform to share their experiences in encountering, raising, and answering new ethical questions as well as engage with the public on these topics. The event featured Northwestern synthetic biologists Danielle Tullman-Ercek, Julius Lucks, and Josh Leonard, as well as Artist-at-Large Dario Robleto and professor of medical education and anthropology Megan Crowley Matoka. Discussions ranged from capturing the ethics of science as art to engage two separate fields and the public, how to best get research to the point of need, and the cultural and ethical impact of organ transplant donation.
See the press release about the event here.
Northwestern hosts 6th International Mammalian Synthetic Biology Workshop
The conference took place from May 17th-19th on Northwestern University’s campus and brought in researchers from around the world with over 150 in attendance. The first ever pre-conference workshop tutorial series took place on Friday, May 17th, providing an open forum for those new to and experienced in mammalian synthetic biology to ask questions to experts about topics such as sensing, cell signaling and differentiation, gene expression, and protein engineering, with professors Julius Lucks, Leonardo Morsut, Tara Deans, and Danielle Tullman-Ercek leading the discussions. Saturday and Sunday featured talks from experts in mammalian synthetic biology such as Linda Griffith, Melody Swartz, Ron Weiss, Martin Fussenegger, and many more. Leonard Lab member Devin Stranford gave a talk on her extracellular vesicle research and its applications to treating HIV. Additionally, Leonard Lab members Joe Muldoon, Taylor Dolberg, Patrick Donahue, Alex Prybutok, Kate Dray, Joseph Draut, Brandon Lim, Vis Kandula, Parth Shah, and Anthony Kang presented posters at the poster session on Saturday evening. Sunday’s talks concluded with a panel discussing the broader ethical implications and impacts of synthetic biology research, featuring Artist-at-Large Dario Robleto, bioethicist Laurie Zoloth, and historians Ben Hurlbut and Gaymon Bennett. Josh was presented with a commemorative plaque for helping organize and host the conference, which will take place by the Centre for Synthetic and Systems Biology at the University of Edinburgh, Scotland.
See the full press release about the conference here.
Anthony Kang awarded HPME Summer Research Grant
Joseph successfully defends his master’s thesis!
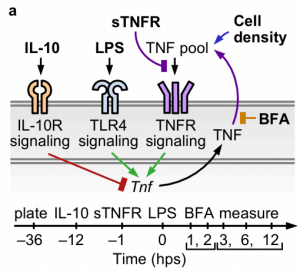
New paper: Macrophages employ quorum licensing to regulate collective activation
We’re thrilled to share our work on macrophage quorum licensing. Congratulations to Joe, Yishan, and our key collaborator Neda Bagheri! This exciting work involved both experimental and computational efforts and a lot of hard work! Check out the published work here and the NU press piece here!
Vis receives top award at Stanford Undergraduate Research Conference
Congratulations to Vis Kandula, who received the “Outstanding Presenter” award at this year’s Stanford Undergraduate Research Conference, which took place from April 5-7. Of the 400 participants that applied from around North America, 80 individuals were accepted to present their research projects ranging from topics about experimental life sciences to quantitative social sciences. Vis was selected as an Outstanding Presenter in the experimental life sciences category.
Vis awarded grant to attend Undergraduate Research Conference
Liz Schauer awarded undergraduate grant
Amy successfully defends her master’s thesis!
Vis Kandula wins CLP undergraduate research award
Amy Hong wins Fletcher Undergraduate Research Prize
Summary of Inaugural Central US Synthetic Biology Workshop
Benji successfully defends his master’s thesis!
Announcing the Inaugural Central US Synthetic Biology Workshop
Northwestern and the Center for Synthetic Biology will host the inaugural Central US Synthetic Biology Workshop on Sep 6-7, 2018. The goal of this event is to create a recurring workshop that brings together synthetic biology researchers and companies from institutions in the Central US. Visit the workshop site and read more here: www.centralsynbio.org
New project funded by NIH’s new synbio-specific mechanism
Our new collaborative project, which is joint with Neda Bagheri, will develop new experimental and computational tools for designing and building robust cell-based therapies. This project was funded in the first round of the NIH’s new synthetic-biology-specific funding mechanisms. Read more here, and a related article released by the Chicago Biomedical Consortium here.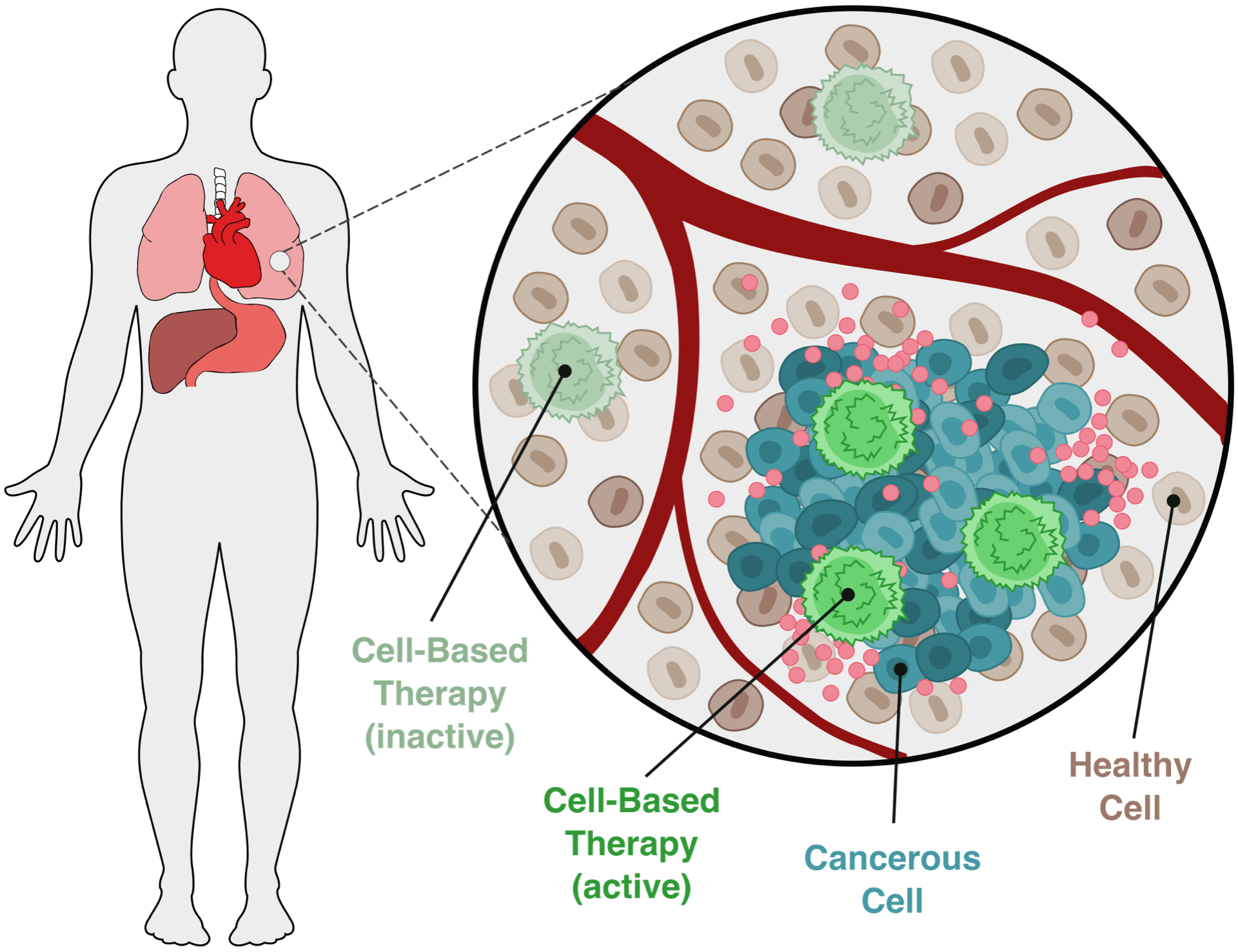
Josh gives a TedX Talk on Synthetic Biology
Check out Josh’s talk, which was given at TEDxYouth@ASFM themed V³: Vision. Voice. Vigor. The event was hosted by the American School Foundation of Monterrey, Mexico.
Read more about the event here, and check out the talk on YouTube.
Devin and Taylor receive top presentation awards at ASGCT
Josh named a Charles Deering McCormick Professor of Teaching Excellence
This is the highest teaching award conferred by Northwestern University. Read more at the two stories below:
https://news.northwestern.edu/stories/2018/may/celebrating-excellence-in-teaching/?linkId=51693991
Lauren earns a summer undergraduate research grant (two, actually!)
Congratulations to Lauren Battaglia, who was selected as a Chemistry of Life Processes Summer Scholar, as well as for a McCormick Summer Research Award!
Ashwin, Amy, and Joseph earn summer research grants
Hailey Edelstein awarded NSF Graduate Research Fellowship
Taylor and Patrick publish News & Views in Nature Chemical Biology
Taylor and Patrick co-authored a commentary on recent progress in engineering cell therapies for cancer, including perspectives on exciting recent innovations in this space. Read more here.
MESA tech for engineering cell therapies highlighted in Abbvie Science Blog
Abbvie senior scientist Mikkel Algire highlights technologies for engineering cell-based therapies for cancer. Read more here.
Anthony publishes a study on ultra-small polymer-caged nano-bins
This collaboration with the Nguyen and Schatz labs decribes the development of highly stable and stimuli/pH-responsive ultra-small polymer-grafted nanobins (usPGNs) as candidate nano-scale drug delivery vehicles. Read more here.
Michelle, Stephen, and Devin publish a chapter on using chromatography to study EV populations
This chapter was published as part of an issue of Methods in Molecular Biology dedicated to the study and therapeutic use of Extracellular RNA. Read more here.
Andrew, Peter, Andrea, and Shreya publish study describing new method for biosensor engineering
This study, published in Protein Engineering, Design, and Selection, reports the BERDI method for engineering novel metabolite-responsive biosensors, for applications ranging from metabolic engineering to fundamental research. Read more here.
Joseph featured in McCormick fall magazine
Joseph’s research is highlighted in an article featuring undergraduate summer research. Read more here.
Joe, Patrick, and Taylor publish a new review on engineered cell therapies in Current Opinion in Biomedical Engineering
This review discusses both recent advances and emerging general principles for using mammalian synthetic biology to build cell-based therapies that are safe and effective. Read more here.
Devin’s Chapter on EV Engineering is published in Advances in Genetics
This chapter summarizes key strategies and challenges associated with studying and harnessing EV-mediated transfer for therapeutic applications, while providing contemporary perspectives on how choices of systems design and analysis method may impact the efficacy of EV-based therapies and the interpretation of EV-based investigations. Read more here.
Kelly profiled in NU Alumni Magazine
Kelly’s thesis research was profiled in a feature in Northwestern’s Alumni Magazine
Josh joins scientific leadership of GP-Write
Josh was selected to join the Scientific Executive Committee of GP-Write, a multinational synthetic biology project dedicated to realizing whole genome synthesis in order to understand, engineer and test living systems.
New SynBio for Cancer Immunotherapy Project
As part of former Vice President Joe Biden’s Cancer Moonshot Initiative, we received an award from NSF to work with fellow synthetic biologists at MIT to develop novel technologies for enabling cancer immunotherapy.
Josh elected to AIChE Leadership Position
Rachel, Kelly, and Joe’s paper on Multiplexing MESA receptors is published in ACS Synthetic Biology
This paper marks the first demonstration that MESA receptors can be multiplexed to function orthogonally in a single cell. We also report here, for the first time, a quantitative model for evaluating and ultimately designing new MESA receptors and new MESA-based technologies. Read more here.
Devin’s paper on targeting EVs to breast cancer is published in Tissue Engineering Part A
Congrats to Devin! Read more here.
Cameron McDonald receives SIGP summer undergraduate research grant!
Cam McDonald will work with Joe Muldoon and Patrick Donahue on a synthetic biology project this summer, supported by the Northwestern Summer Internship Grant Program (SIGP). Congratulations, Cam!
Joseph Draut receives McCormick summer undergraduate research award!
Joseph Draut will work with Patrick Donahue on a synthetic biology project this summer, supported by the McCormick School of Engineering and Applied Science. Congratulations, Joseph!
Amy Hong receives summer undergraduate research grant!
Amy Hong will work with Joe Muldoon on a computational synthetic biology project this summer, supported by the Northwestern Office of Undergraduate Research. Congratulations, Amy!
NU Hosts EBRC Retreat!

Held at the Norris University Center on March 24-25, the Engineering Biology Research Consortium brought together nearly 170 faculty, researchers, and students from universities across the country to discuss current challenges and opportunities in synthetic biology.
Taylor receives Poster Presentation Recognition at EBRC Retreat
Congratulations to Taylor, who received an honorable mention / runner up amongst a very competitive field of poster presenters at this past weekend’s EBRC retreat, hosted right here at Northwestern by the Center for Synthetic Biology!
Kelly’s NCB paper highlighted in SBE (Society for Biological Engineering) Newsletter
Nature Chemical Biology publishes a News & Views article about Kelly’s NCB Paper
Synthetic biology: Sensing with modular receptors
Matthew Brenner, Jang Hwan Cho and Wilson W Wong
doi:10.1038/nchembio.2290
Sensing and responding to diverse extracellular signals is a crucial aspect of cellular decision-making that is currently lacking in the synthetic biology toolkit. The development of modular receptor platforms allows for the rewiring of cellular input-output relationships.
Kelly’s Nature Chemical Biology paper covered by GEN
Genetic Engineering and Biotechnology News ran the following press article:
Younger et. al ACS Synthetic Biology piece featured by McCormick Press
Josh interviewed by WTTW for Kelly’s NCB paper
Schwarz et al. Nature Chemical Biology paper featured in NU press release
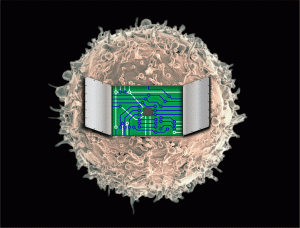 (click image to see animation!)
(click image to see animation!)
Image by Joshua Leonard and Kelly Schwarz, Northwestern University. Cell image by NIAID/NIH, used and modified under Creative Commons 2.0 license.
Seminal MESA paper published in Nature Chemical Biology
Congrats to Kelly, Nickey, and Taylor, whose paper was published today in Nature Chemical Biology! This paper is currently spotlighted on the homepage of NCB: http://www.nature.com/nchembio/index.html
Leonard lab research featured
Our research was featured in an interview with Josh in Cancer Cytopathology, for a special feature on synthetic biology technologies for treating cancer (Nov 2016) [link]
Andrew’s new biosensor engineering paper, co-authored with former undergraduate mentees Neil and Austin, was published in ACS Synthetic Biology
Congratulations to Andy and Andrew!
Andy and Andrew’s paper was accepted for publication in ACS Synthetic Biology (a collaboration with our partners at the University of Warwick)
Congratulations to Joe!
Joseph Muldoon received the 2015-2016 IBiS Teaching Assistant Excellence Award
Congratulations to Kelly!
Kelly Schwarz was selected as runner-up for this years Distinguished Graduate Researcher Award from the Department of Chemical and Biological Engineering.
Congratulations to Michelle!
Michelle’s collaborative paper with Carol Hall at NC State was published in the Journal of Computational Chemistry.
Josh takes on leadership role in BTP
Josh is now the Co-Director of Northwestern’s NIH-funded Predoctoral Biotechnology Training Program
Congratulations to Josh!
Josh was selected to give the annual graduate student invitational seminar to the Department of Biomedical Engineering at the University of Virginia.
Congratulations to Yishan, Michelle, and Brianne!
Yishan, Michelle, and Brianne’s paper was on macrophage decision making in response to IL-10 was accepted for publication in Innate Immunity.
Congratulations to Josh!
As part of President Obama and Vice President Biden’s Cancer Moonshot Initiative, Josh will be an invited conferee at a meeting focused on “Systems and Synthetic Biology for Designing Rational Cancer Immunotherapies” in October.
Congratulations to Andrew!
Congratulations to Andrew on being appointed to the Biotechnology Training Program!
Science Magazine technology feature
Josh is interviewed and Michelle’s research is featured in a Science Magazine technology feature on extracellular vesicles. Read more here:
Congratulations to Dr Michelle Hung!
Congratulations to Neil
Congratulations to Neil who was awarded the second place Mickelson Prize at this year’s McCormick Graduation Convocation!
Congratulations to Andrew and Jeci!
Patrick wins an NRSA!
Congratulations to Patrick who was just awarded the Ruth L. Kirschstein National Research Service Award (NRSA) fellowship from NIH!
Congratulations to Praneet!
Congratulations to Praneet who was just awarded a Weinberg College Summer Research Grant!
Congratulations to Michelle!
Congratulations to Michelle who recently published her 2nd first author publication in the Journal of Extracellular Vesicles! Read it here
Congratulations to Andrea and Imran!
Andrea and Imran were selected to receive a Summer Undergraduate Research Grant (URG)! Congratulations!
Congratulations to Kelly: March Madness Bracket Champion!
Congratulations to Kelly Schwarz who won this years Leonard Lab Bracket Challenge!
Welcome Andrea, Imran, and Praneet to the lab!
Welcome to our new undergraduate researchers Andrea Shepard, Imran Khan, and Praneet Polineni!
Devin wins an NSF GRFP!
Congratulations to Devin who won a NSF fellowship!
Neil Dalvie accepts MIT’s offer for graduate studies
Undergraduate researcher Neil Dalvie has accepted MIT’s offer for graduate studies for the anticipated degree of Ph.D.
Congratulations and good luck!
Data as Art
Northwestern’s Center for Synthetic Biology Opens!
Congratulations to Michelle!
Michelle Hung won a meritorious travel grant from the American Society of Gene & Cell Therapy (ASGCT)!
Welcome Pear to the lab!
Welcome to our new undergraduate researcher Pear Dhiantravan!
Welcome Shreya to the lab!
Welcome to our new undergraduate researcher Shreya Udani!
Welcome Devin and Peter to the lab!
Welcome to our two new Chemical Engineering PhD students Devin Stranford and Peter Su!
Josh Leonard gets promoted!
Congratulations to Josh who got officially promoted to Associate Professor with tenure!
Congratulations!
Andy Scarpelli successfully defended his thesis!
Welcome Austin to the lab!
Austin Rottinghuas joins our lab for a summer research project from Northwestern’s Summer Research Opportunity Program (SROP)
Congratulations!
Rachel Dudek successfully defended her thesis!
Public recognition
Wells et al. paper highlighted in Northwestern McCormick Engineering News! Read More
Public recognition
Hung et al. was one of the most viewed JBC articles published in March and April! Check out the article! Pubmed
Ph.D. student award!
Congratulations to Michelle Hung for being selected for the Best Mentor Award from Niles West High School
Undergraduate award!
Congratulations to Neil Dalvie for being awarded the Chemistry of Life Processes Summer Scholar Award!
PH.D. student award!
Congratulations to Taylor Dolberg for winning the NDSEG graduate research fellowship!
Ph.D. student award!
Congratulations to Taylor Dolberg for winning the NSF graduate research fellowship!
Ph.D. student award!
Congratulations to Joe Muldoon for wining the NSF Honorable Mention!
New publication!
Congratulations to Ragan Pitner and Andy Scarpelli for their recent paper in ACS Synthetic Biology! Get it here ACS
New publication!
Congratulations to Michelle Hung for her recent paper in JBC! Pubmed
New publication!
Congratulations to Danny Wells and Yishan Chuang for their recent paper in PLOS Computational Biology. Read it here Pubmed
New publication!
Congratulations to Nichole Daringer and Kelly Schwarz for their recent paper in JBC! Get it here JBC
Undergraduate award!
Congratulations to Naomi Lisse for winning an Undergraduate Research Grant!
Honor
Michelle Hung invited to speak at the IBiS mini-symposium!
Undergraduate award!
Congratulations to Neil Dalvie for winning an Undergraduate Research Grant!
Welcome Taylor to the lab!
Welcome to our new Chemical Engineering Ph.D. student Taylor Dolberg to the lab!
New publication!
Congratulations to Rachel Dudek, Yishan Chuang, and Josh Leonard for publishing in Advances in Experimental Medicine and Biology! Read it here Pubmed
Special publication!
The Leonard Lab features prominently, including the cover art (by Justin Muir), in the current ACS Synthetic Biology issue! ACS Synthetic Biology Special Issue: Viewpoint by Josh Leonard, Podcast interview with Josh Leonard, Daringer et al!
Honor
Dr. Leonard and Dr. Bagheri receive a Cornew Innovation Award from CLP
@Addgene tweets about the lab!
@Addgene Tweets about the Modular Extracellular Sensor Architecture plasmids now available from the Leonard Lab #SynBio
Congratulations!
Congratulations to Kelly Schwarz for passing her qualifying exam!
Congratulations!
Congratulations to Andrew Younger for being awarded a EPA STAR graduate fellowship!
Congratulations!
Congratulations to Andrew Younger for being awarded the Neil Welker IBiS Teaching Assistant Excellence Award!
Congratulations!
Congratulations to Kelly Schwarz for being awarded the ChBE Departmental Teaching Assistant award!
Welcome Patrick to the lab!
Welcome to our new MD/Ph.D. student Patrick Donahue to the Lab!
Congratulations!
Congratulations to Joseph Muldoon on being appointed to the Biotechnology cluster!
New publication!
Congratulations to Michelle Hung on her first first author research article! Read More
Public recognition
Daringer et al. paper highlighted in the Society for Biological Engineering Connections E-Newsletter Read More
Congratulations!
Congratulations to Nichole Daringer for successfully defending her thesis!
Congratulations
Congratulations to Andrew Younger for passing his qualifying exam!
Public recognition
Research featured on Northwestern’s Office for Research Discovery website Read More
Congratulations!
Congratulations to Yishan Chuang for successfully defending her thesis!
Welcome Joe to the lab!
Welcome to our new IBiS Ph.D. student Joseph Muldoon to the lab!
Honor
Congratulations to Andrew Younger for being selected for the Best Mentor Award from Niles West High School
Honor
Congratulations to Michelle Hung for being awarded the Excellence in Research Award from the American Society of Cell and Gene Therapy
Congratulations!
Congratulations to Danny Wells for being selected for a Marine Biology Lab summer course
Science Fair Results!
Niles West High School students, mentored by Andrew Younger, get invited to the Illinois State Science fair and the Intel International Science and Engineering fair in Los Angeles. Good Luck!
Public recognition
NU Synthetic Biology highlighted on university home page Read More
Synthetic biology workshop
Dr. Leonard and Northwestern University co-host UK-US workshop on Synthetic Biology